Substrate-Aware Zero-Shot Predictors for Non-Native Enzyme Activities
Abstract
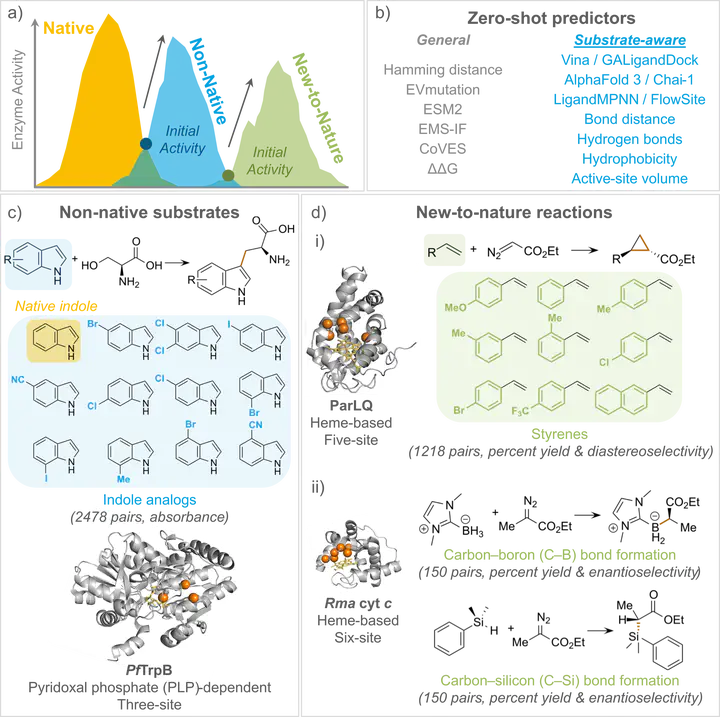
Enzymes can be engineered to catalyze reactions with non-native substrates or even perform entirely new reactions unknown in nature. However, developing such novel activities through wet-lab engineering is time- and resource-intensive. By estimating enzyme activity without new experimental data, zero-shot (ZS) predictors can accelerate enzyme engineering. While ZS predictors have been demonstrated in various contexts, they have yet to be evaluated on non-native sub- strates and new-to-nature chemistry. Critically, many existing predictors do not explicitly encode substrate or transition-state properties, which we propose are essential for predicting new-to-nature chemistry. Here, we systematically studied two types of mechanistically distinct enzymes using 16 ZS predictors—including six general and ten substrate-aware scores derived from generative modeling, molecular docking, and active-site heuristics. We curated new experimental and literature mutation datasets spanning 11 non-native substrates and three new-to- nature reactions with 11 additional substrates. The six general ZS predictors could generalize to non-native substrates, but failed for new-to-nature chemistries. In contrast, certain substrate-aware approaches could predict new-to-nature chemistries, with AlphaFold 3’s chain-predicted aligned error being the most pre- dictive of both activity and stereoselectivity. A weighted ensemble of AlphaFold 3 and EVmutation scores generalized to all chemistries that we tested. Our find- ings highlight the potential of ZS predictors to accelerate enzyme engineering, advancing the expansion of the chemical universe beyond nature’s repertoire.